
07 Oct Data Management at Scale: Lessons from Curating 350,000 Microbial Strains
By Adam G. Newman, Jonathan Vadnal, Deeya Burman – Ginkgo Bioworks
Ginkgo Bioworks’ 350,000+ Strains Microbial Library
Ginkgo Bioworks is a biotechnology company based in Boston focused on making biology easier to engineer. By leveraging our team’s expertise and cutting-edge tools for high-throughput R&D, our customers address major global challenges with biotechnology. For customers seeking novel starting points for new biological products, we offer access to the Ginkgo Bioworks Microbial Library: a collection of over 350,000 microbial strains curated from a variety of sources. The collection contains significant genetic diversity, making our strains well-suited for many biotechnology applications ranging from unique proteins and enzymes, expression systems, and strain engineering. In particular, the collection has nearly 2 million biosynthetic gene clusters encoding natural products that could have benefits to human and environmental health.
Our library has a global provenance with strains originating from 25 countries across all continents except Antarctica. Originally curated with the intent of studying their agronomic benefits, strains in the library were sourced from soil, plants, insects and other animals making them particularly suitable for agricultural uses. For example, many strains in our collection have been found to produce novel natural products to control fungal pathogens while others make N, P, and K more readily available to crops.
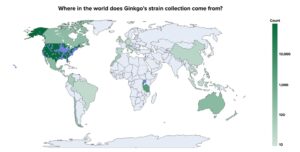
Ginkgo’s Strain Collection Map | Image credit: Ginkgo Bioworks
Physical Infrastructure: When Freezers Aren’t Enough
Through the years, the team at Ginkgo has learned many lessons for how to not only manage a large collection but also to render it functional at scale. Two critical components are physical and digital infrastructure.
As a collection scales, the physical infrastructure supporting that collection must also scale. While maintaining samples in a conventional -80°C freezer might be sustainable for a small collection, such a storage system for a large collection begins to sacrifice the integrity of all samples. Freezers can fail. Samples can be misplaced or lost. Opening a freezer can affect temperature control for thousands of strains. Integration of automated storage and control systems for a large collection not only serves to maintain the integrity of the collection, but also frees up researchers to do what they do best: great science!
Meet The Biostore: An Automated Storage Solution
We house our library in the Biostore at Ginkgo Bioworks: a state-of-the-art, automated microbial storage system located at the West Sacramento, CA facility. As the main repository for the company’s extensive strain collection, the Biostore supports both research and archival needs. The Biostore is equipped with an Azenta (formerly Brooks) BioStore II Twinbank, an automated ultra-low temperature freezer maintained at -80°C. Most importantly, this system is designed for automated storage, retrieval, and inventory management of microbial strains, guaranteeing high sample integrity and secure, auditable, and on-demand access.
In its current configuration, the Biostore allows for storage of up to 1.2 million individual samples. Samples can be stored in a variety of tube sizes, from 0.3 μL 2D barcoded tubes up to 1.8 mL cryovials. While the Biostore is primarily configured for tube-based storage, additional engineering could allow storage of other formats, such as microtiter plates. The storage environment is highly controlled: all freezers are equipped with CO2 backup, environmental monitoring, and building management system (BMS) integration. Access is restricted to trained personnel to ensure biosafety and sample security. Samples are managed using 2D barcoded tubes for precise tracking and to minimize handling errors.

Ginkgo’s Biostore | Image credit: Ginkgo Bioworks
Digital Infrastructure: Tracking Every Strain
With large collections, it is critical to make sure that your physical infrastructure is backed up with digital infrastructure. At Ginkgo, we have built a custom database to store critical metadata on each strain in our collection. It is critical that each strain receives an unique and immutable identifier, allowing us to track strains throughout their lifecycle. All other data associated with the strain can then be linked to that identifier, giving the organization a complete digital audit trail on a strain. In this way, as new data is collected on a strain (for example, a genome assembly is created) or as the information on the strain comes into higher resolution (for example, the taxonomic classification is updated), researchers can amend our database without creating confusion or data collisions. Furthermore, the Biostore is fully integrated with the Ginkgo’s Laboratory Information Management System (LIMS), providing a complete digital view for every strain and high confidence in physical sample location and status. With digital copies for each of our strains, our scientists can source strains from our library that meet the commercial needs of our customers with an eye towards freedom-to-operate.
Genomic Analysis: Understanding Genetic Diversity
In addition to metadata, Ginkgo has sequenced nearly two thirds of the strains in our library, attaching draft genomes to more than 200,000 microbial strains. In a strain collection that spans 1,900+ unique species, genetic overlap is expected, but minor differences are not taken for granted. Our experts in ag biologicals have observed firsthand how even small genetic differences between strains within the same species can have profound phenotypic effects. Different species within the same genus can show significant differences in traits like nitrogen fixation and ammonia secretion.
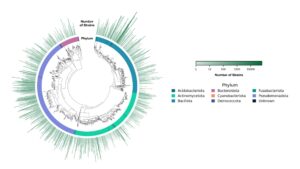
Ginkgo’s Strain Tree | Image credit: Ginkgo Bioworks
The OMG Platform: Processing Genomes at Scale
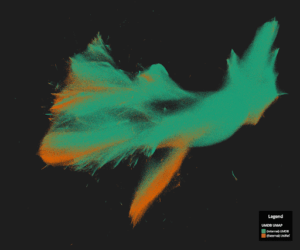
UMDB Protein Map | Image credit: Ginkgo Bioworks
To understand these genetic features efficiently, Ginkgo has built a custom genome assembly and annotation pipeline, capable of processing 100,000s of microbial genomes in a matter of weeks: the One Microbial Genome (OMG) platform. Not only does OMG perform standard annotations like gene prediction and taxonomic classification, but it also provides custom annotations including agricultural-specific functional pathway identification and antibiotic resistance mechanism determination. These genomic data are stored in a custom database, OMGDB, allowing researchers to effortlessly interrogate the collection on a variety of sequence- and annotation-based queries.
Custom Application Programming Interfaces (APIs) grant researchers access to OMGDB programmatically, allowing them to perform both standard and bespoke bioinformatic analysis on strains in the collection at scale. These data also feed into Ginkgo’s Unified Metagenomic Database (UMDB), which comprises publicly available and proprietary gene sequences. To date, the company has 3.5 billion unique protein sequences in UMDB with 84% (2.97 billion) of these sequences represented solely in Ginkgo’s databases.
Bringing It All Together: An Integrated Approach
Managing a microbial library on the scale of Ginkgo’s 350,000+ strains necessitates a robust combination of physical, digital, and analytical infrastructure. From the automated Biostore ensuring sample integrity to comprehensive digital tracking and advanced genomic analysis platforms, each component is critical for efficient operation. This integrated approach allows Ginkgo to effectively leverage our vast collection, unlocking its full value for diverse biotechnology applications. By embracing these lessons, Ginkgo Bioworks continues to push the boundaries of biological engineering, making biology easier to engineer for the benefit of all.
To learn more about Ginkgo Bioworks, the Ginkgo Bioworks Microbial Library, or to inquire about Ginkgo Bioworks services, follow us on LinkedIn at Ginkgo Agriculture.
